Research
From genes to geometry: how mechanical interactions between tissue layers sculpt visceral organs
Geometry and mechanics often provide a powerful lens for understanding the mechanisms governing material behavior. This perspective extends to biological systems, opening a window into the mechanisms of tissue shape change during embryonic development. In development, organ geometry and tissue mechanics interact with genetic patterning. We address the dynamic and mechanical processes by which biology sculpts complex organ shapes, unraveling the collective cell behaviors and tissue layer interactions that link genes to geometry. Across a broad class of organs including the gut and heart, sheets of nascent muscle cells ensheath underlying epithelial layers to collaboratively guide morphogenesis. We aim to quantitatively decode the supracellular mechanical activity composing these organs’ shape change. This effort invokes advanced microscopy techniques for whole-organ live imaging, genetic & optogenetic perturbations, computational image analysis, differential geometry, and analytic approaches from physics. We leverage the Drosophila and zebrafish embryos as living laboratories.
Multilayer tubulogenesis
A common motif in biology is the arrangement of cells into tube-like sheets. How do coupled mesodermal and endodermal cell layers collectively pattern mechanical forces to build tube-like organs? The digestive organs of the embryonic fly enable us to probe the dynamic processes of multilayer tube formation at cellular and subcellular resolution in vivo — through multiple motifs of tubulogenesis, including multilayer lumen entrapment (left) and multilayer tissue budding (right).

Chiral organ coiling
Organ looping is often assumed to be a process of differential growth — where patterned cell division or area changes lead to bending in 3D. However, some organs coil into looped shapes that break left-right symmetry without any cell divisions and even without asymmetrically patterned morphogens. This speaks to a mechanical contribution at the core of how biology breaks left-right symmetry. We are investigating the cell-intrinsic behaviors that drive chiral (LR asymmetric) organ shapes. How do in-plane tissue deformations such as twist/shear couple to 3D organ coiling, and how are these deformations controlled? Emerging notions of nonreciprocal interactions and odd viscoelasticity in soft matter physics are likely to find ready application in this biological context.
Shape change and mechanical function
The vertebrate heart develops into a coil of chambers while establishing functional, stereotyped mechanical activity needed for blood circulation. We are developing tools to measure the shape change across developmental time in this beating organ using zebrafish as a model system. Our goal is to understand the interactions between cell-scale and organ-scale geometric transformations, as well as the interactions between beating kinematics with shape changes.
Developmental trajectory selection
How do organs select their trajectories in shape space, and what drives morphogenetic robustness? Taking a quantitative approach to parameterizing organ shape and developmental trajectories, we are particularly interested in understanding the ‘bistability’ of chiral shape transformations and their relationship to cellular behavior.

Stay tuned for updates on emerging projects!
PAST PROJECTS (pre-Mitchell Lab)
Folding morphogenesis of a multilayer organ
Visceral organs are composed of multiple tissue layers that interact both mechanically and chemically. During morphogenesis, the initial geometry of visceral organs undergoes a sequence of folding, adopting complex shapes that are vital for function. How do coupled tissue layers choreograph organ shape transformations during embryonic development? Understanding the dynamics of visceral organs has been a formidable challenge, due to both technical and conceptual challenges. We bring advanced microscopy, computational, and theoretical approaches to address how dynamic cellular behaviors generate active stresses to achieve stereotyped, complex shape change in 3D.


We showed how coupled tissue layers generate complex shape transformations in the fly midgut. We find that calcium mediates the contraction of muscle cells in the outer layer, triggering cellular shape transformations in the endodermal inner layer. These shape transformations give rise at the tissue scale to a convergent extension motif that couples in-plane deformations to out-of-plane motion.
publication: N. P. Mitchell, D. J. Cislo, S. Shankar, Y. Lin, B. I. Shraiman, S. J. Streichan, “Visceral organ morphogenesis via calcium-patterned muscle contractions.” eLife 11:e77355 (2022)
Methods for building quantitative models of morphogenesis
Capturing the interplay between in-plane and out-of-plane tissue shape change during development requires following the tissue surface as it deforms and integrating cell-scale motions into collective, tissue-scale deformations. We automatically extract whole-organ measures of deformation from complex, dynamic tissue surfaces in 3D. This enables whole-organ tracking, quantification of tissue deformation and cell intercalations, tissue anisotropy, and more.

publication: N. P. Mitchell*, D. J. Cislo*. “TubULAR: Tracking deformations of dynamic tissues via constrained maps.” Nature Methods (2023) [Full text, biorxiv]
see also: A. M. Gabbert, J. A. Mondo, J. P. Campanale, N. P. Mitchell, A. Myers, S. J. Streichan, N. Miolane, D. J. Montell, “Septins regulate border cell shape and surface geometry downstream of Rho.” Developmental Cell (2023) [biorxiv, Full text]
A Morphodynamic Atlas for Embryonic Development
Mapping embryonic development at cellular resolution across the whole Drosophila embryo surface enabled a large, unified atlas of gene expression and tissue deformation. This atlas features fly embryos from 0-24 hrs of development, including whole-animal datasets of organ development at high temporal resolution.
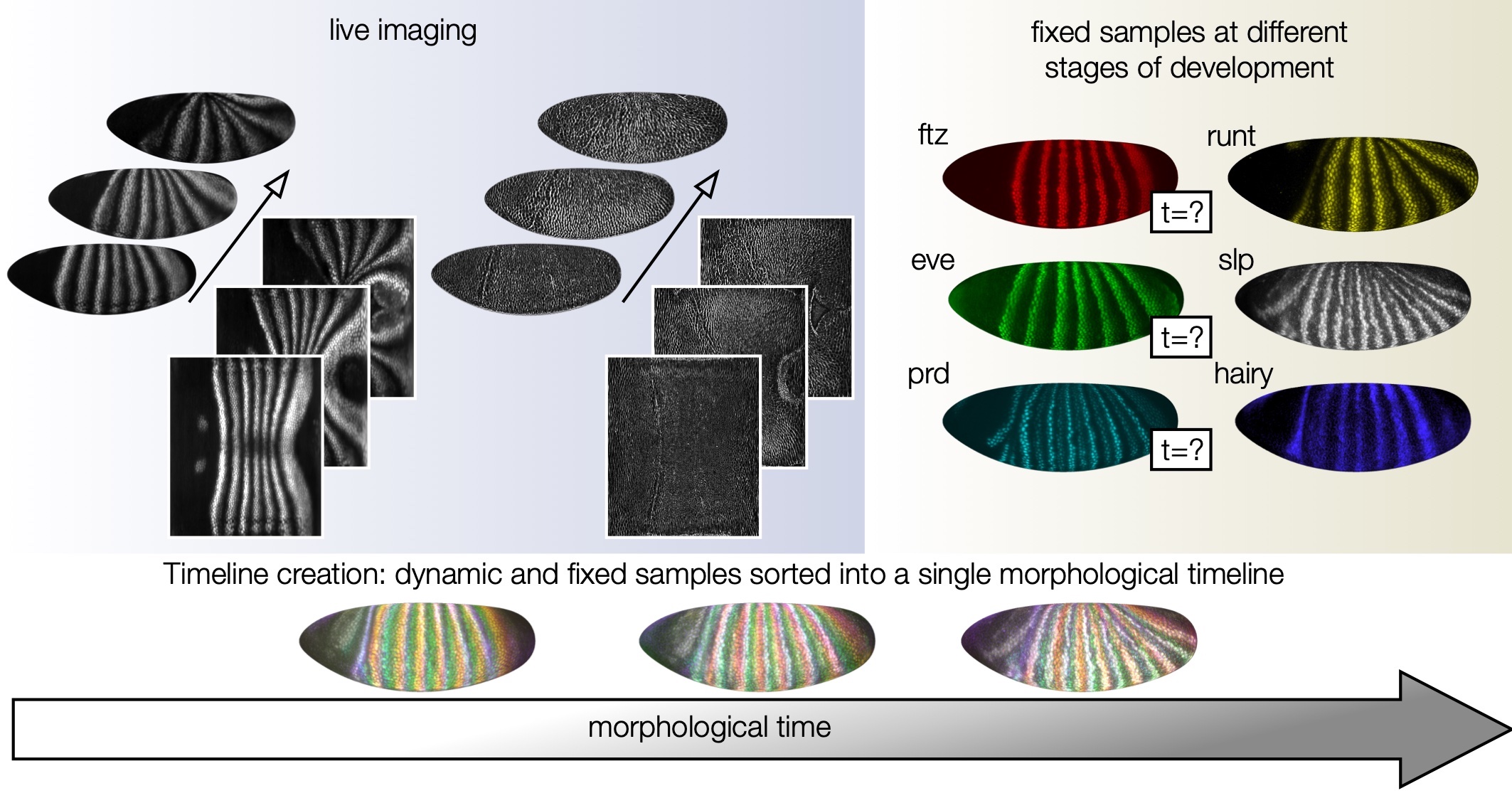
publication: N. P. Mitchell*, M. F. Levebvre*, V. Jain-Sharma*, N. Claussen, M. K. Raich, H. J. Gustafson, A. R. Bausch, S. J. Streichan. “Morphodynamic atlas of Drosophila development.” [biorxiv] (2022)
Self-organized tissue flow in embryonic gastrulation
By careful measurements of the tissue deformations during early embryogenesis in Drosophila, we find that anisotropic myosin patterns in the swirling tissues exhibit remarkable self-similarity over time –– far more than would be possible if the tissue was advecting these patterns. This work and ongoing collaborations with Sebastian Streichan’s lab attempt to unravel the different contributions to mechanical deformations in the early embryo.
publication: M. F. Lefebvre*, N. H. Claussen*, N. P. Mitchell, H. J. Gustafson, S. J Streichan. “Geometric control of Myosin-II orientation during axis elongation.” eLife 2023 [biorxiv]
Geometry, topology, & dynamics in biological matter
How do cytoskeletal components self-organize to generate intricate structures? In collaborations with the Dogic Lab (UCSB), we studied reconstituted gels made of microtubules and the molecular motor kinesin-4 and self-assembled membranes composed of viruses. In each study, we tuned the emergent behavior of these composite materials, controlling their geometry and/or the topology of the aggregate structure.
publication: B. Lemma, N. P. Mitchell, R. Subramanian, D. J. Needleman, Z. Dogic. “Active microphase separation in mixtures of microtubules and tip-accumulating molecular motors.” Physical Review X (2022) [arxiv]
publication: A. Khanra, L. L. Jia, N. P. Mitchell, A. Balchunas, R. A. Pelcovits, T. R. Powers, Z. Dogic, P. Sharma. “Controlling the shape and topology of two-component colloidal membranes.” PNAS 2022 [arxiv]
For summaries of other past projects, see here.
